3.7 Nonlinear Regression - Prediction Plot, Poly
install.packages("splines")
library(MASS)
library(splines)
library(ISLR)
library(tidyverse)
set.seed(45)
Prediction plot
#predictionplot
#Create a function called pred_plot() that takes as input an lm object
pred_plot <- function(model) {
# First create predictions for all values of lstat
x_pred <- seq(min(Boston$lstat), max(Boston$lstat), length.out = 500)
y_pred <- predict(model, newdata = tibble(lstat = x_pred))
# Create a ggplot object with a line based on those predictions
Boston %>%
ggplot(aes(x = lstat, y = medv)) +
geom_point() +
geom_line(data = tibble(lstat = x_pred, medv = y_pred), size = 1, col = "blue") +
theme_minimal()
}
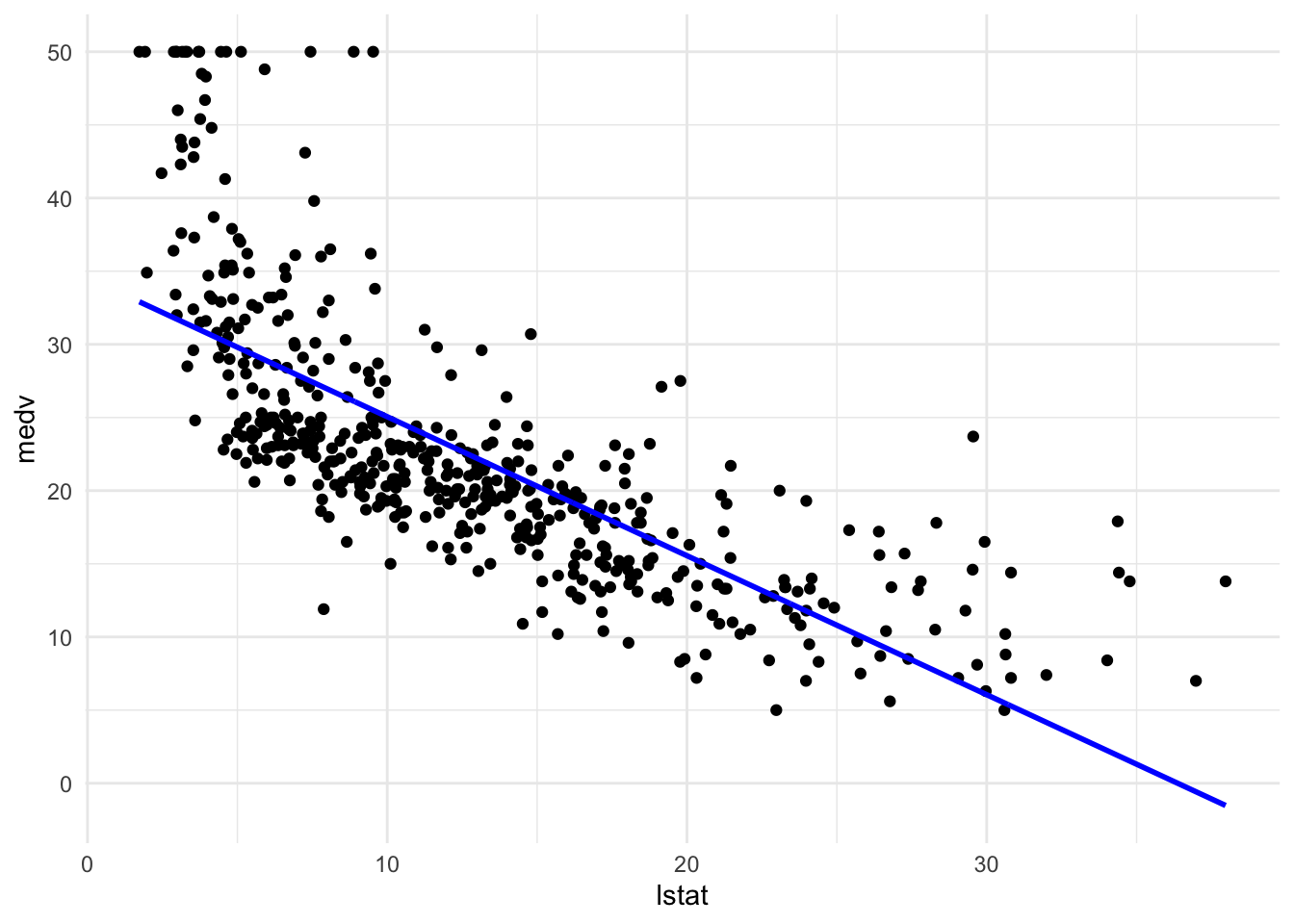
#Create a linear regression object called lin_mod which models medv as a function of lstat
lin_mod <- lm(medv ~ lstat, data = Boston)
pred_plot(lin_mod)
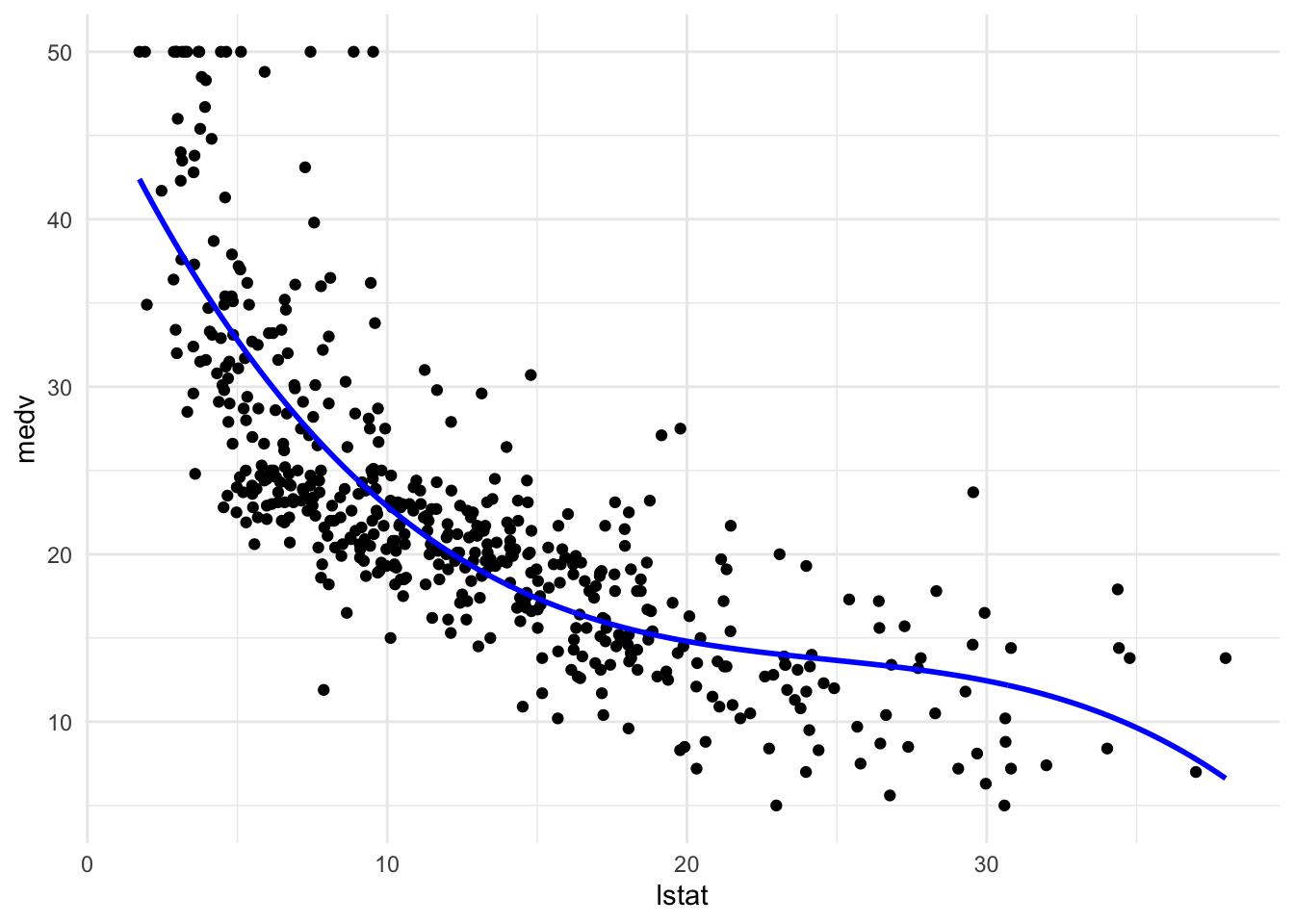
Polynomial regression
#polynomial #polynomialregression
pn3_mod <- lm(medv ~ lstat + I(lstat^2) + I(lstat^3), data = Boston)
pred_plot(pn3_mod)
#适合想要有更多控制的情况,因为可以只选一次方和三次方,而不选二次方
pn3_mod2 <- lm(medv ~ poly(lstat, 3, raw = TRUE), data = Boston)
pred_plot(pn3_mod2)
#poly()适合想要更简洁的情况,因为不能自主设定选几个不同的次方
Piecewise regression
#piecewise #piecewiseregression
适合情况:relationship between variables might change abruptly at a certain threshold,比如农作物低于某个气温,产量有很大变化
#设定用于分成几个piece的数值
#Create a model called pw2_mod with one predictor: I(lstat <= median(lstat)).
#Create a pred_plot with this model.
#Use the coefficients in coef(pw2_mod) to find out what the predicted value for a low-lstat neighbourhood is.
pw2_mod <- lm(medv ~ I(lstat <= median(lstat)), data = Boston)
pred_plot(pw2_mod)
coef(pw2_mod)
## (Intercept) I(lstat <= median(lstat))TRUE
## 16.67747 11.71067
# the predicted value for low-lstat neighbourhoods is 16.68 + 11.71 = 28.39
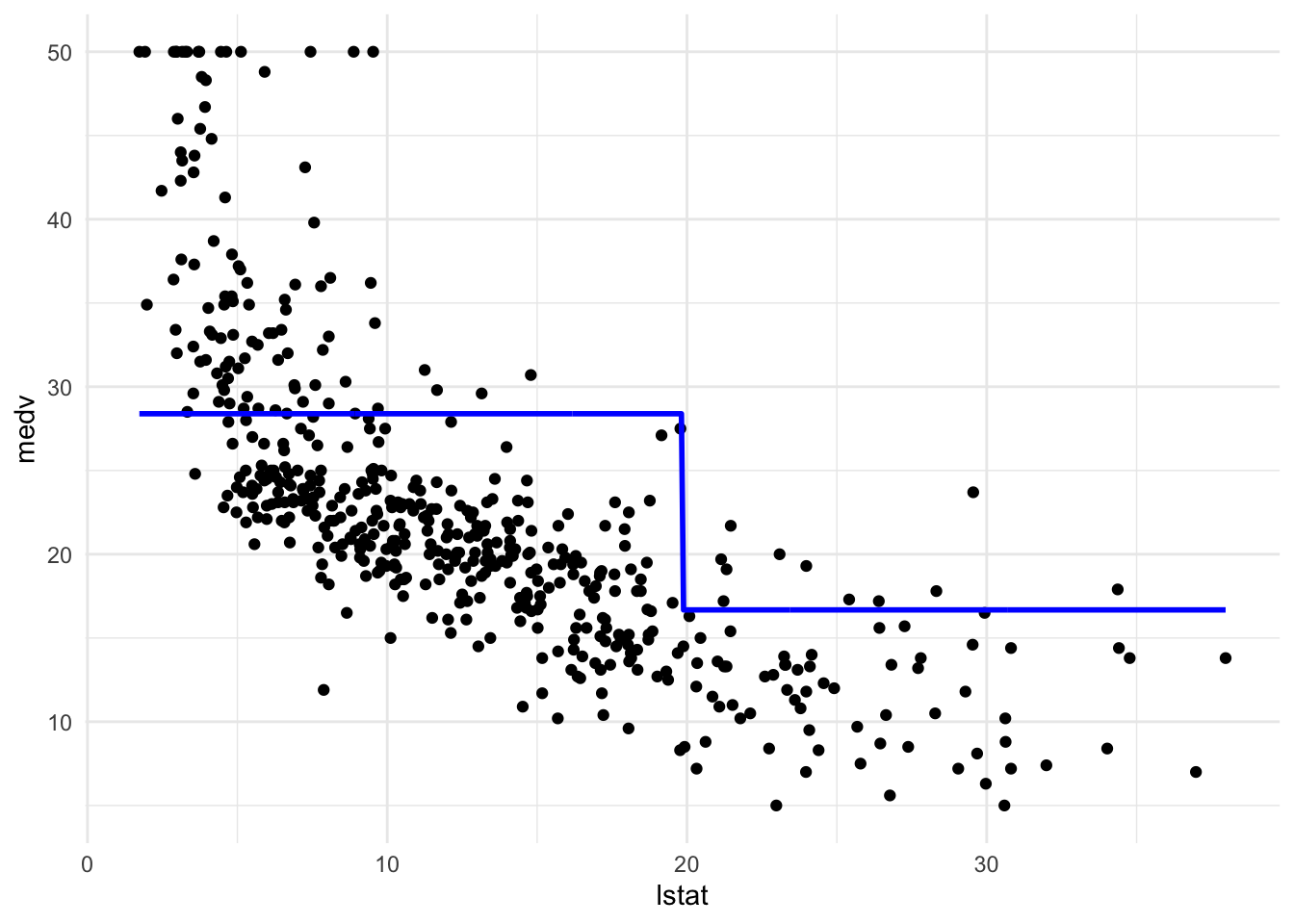
#设定分成n个(space相同的)piece
pw5_mod <- lm(medv ~ cut(lstat, 5), data = Boston)
pred_plot(pw5_mod)
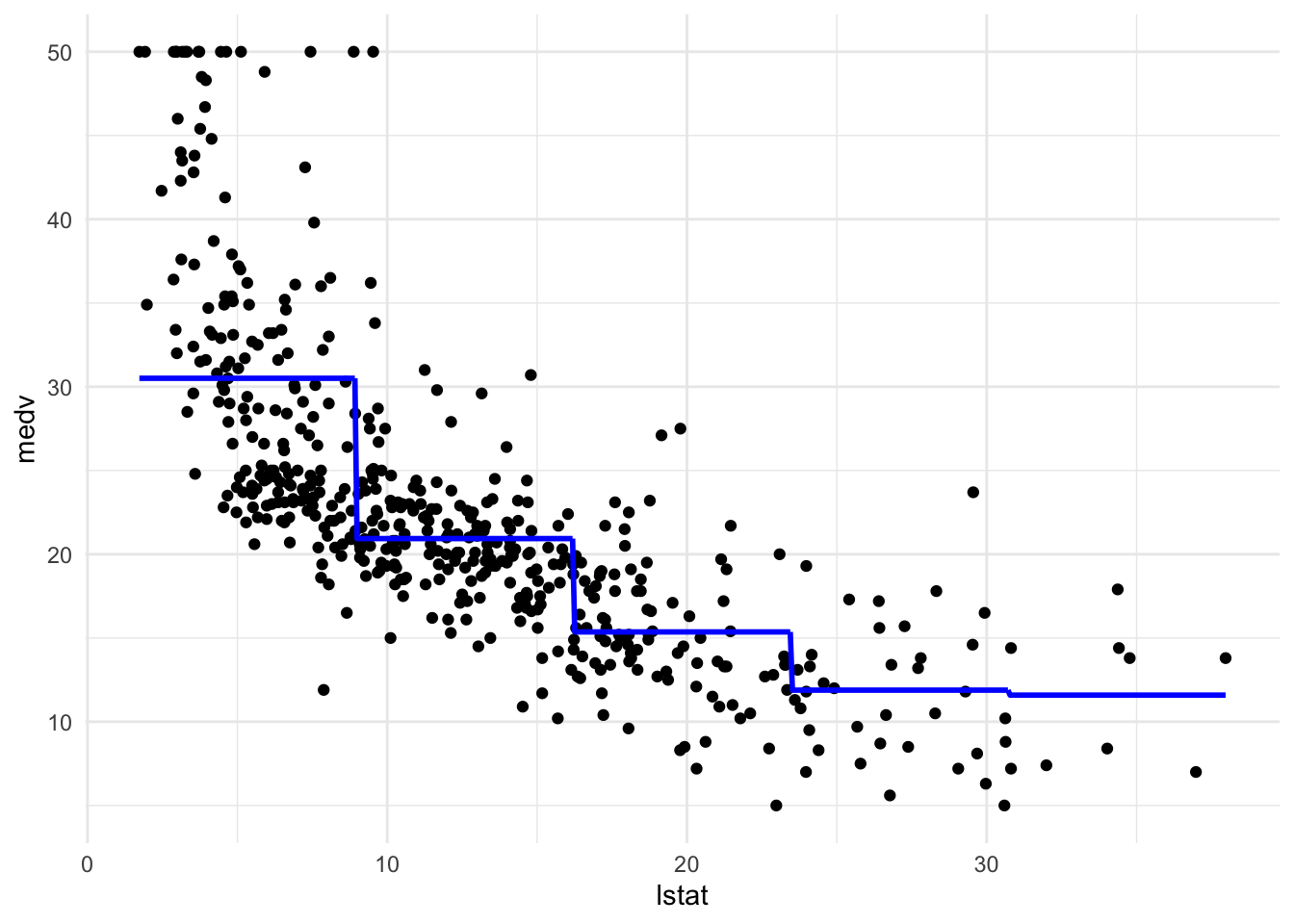
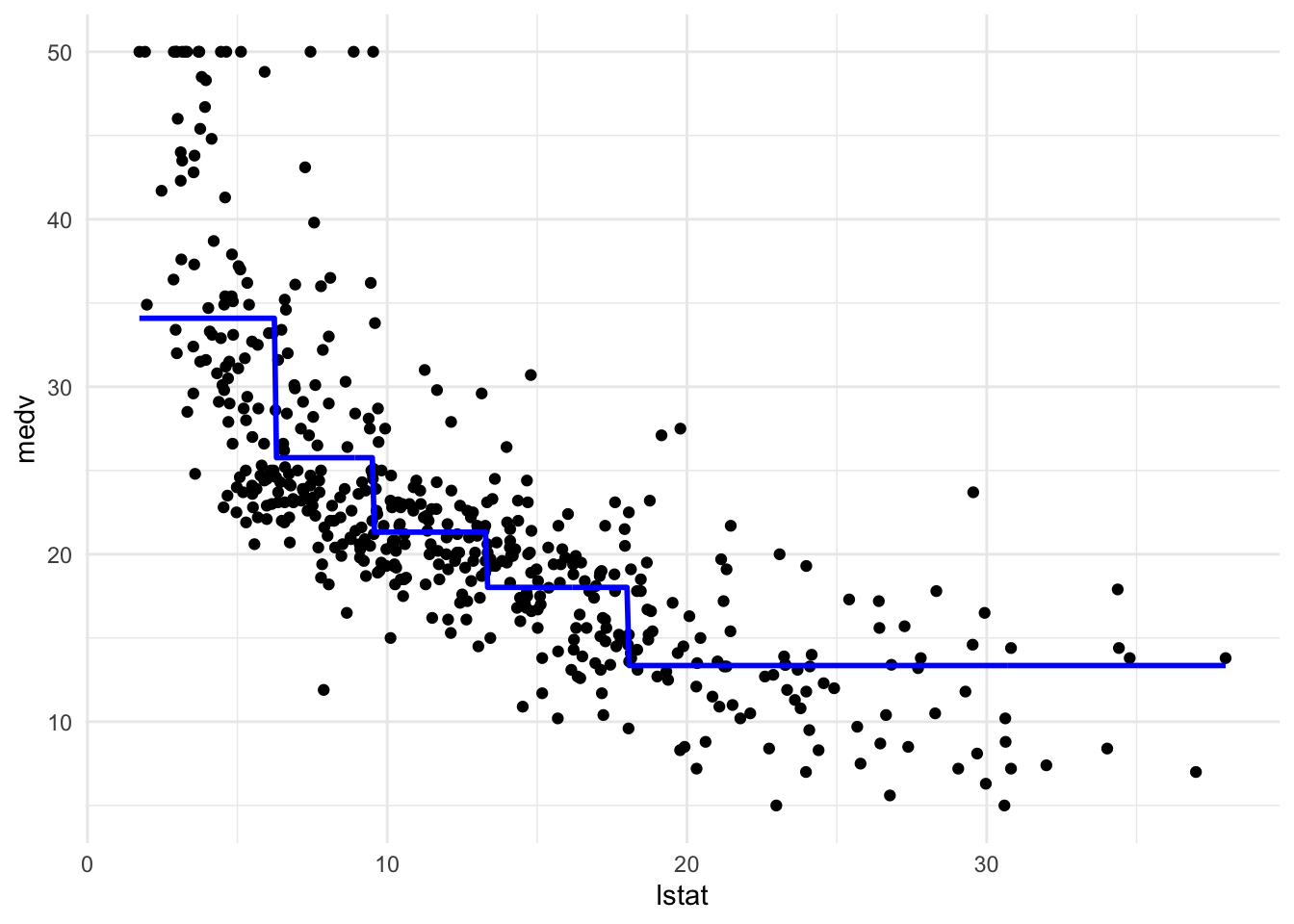
#设定分成n个(datapoint相同的)piece
brks <- c(-Inf, quantile(Boston$lstat, probs = c(.2, .4, .6, .8)), Inf)
pwq_mod <- lm(medv ~ cut(lstat, brks), data = Boston)
pred_plot(pwq_mod)
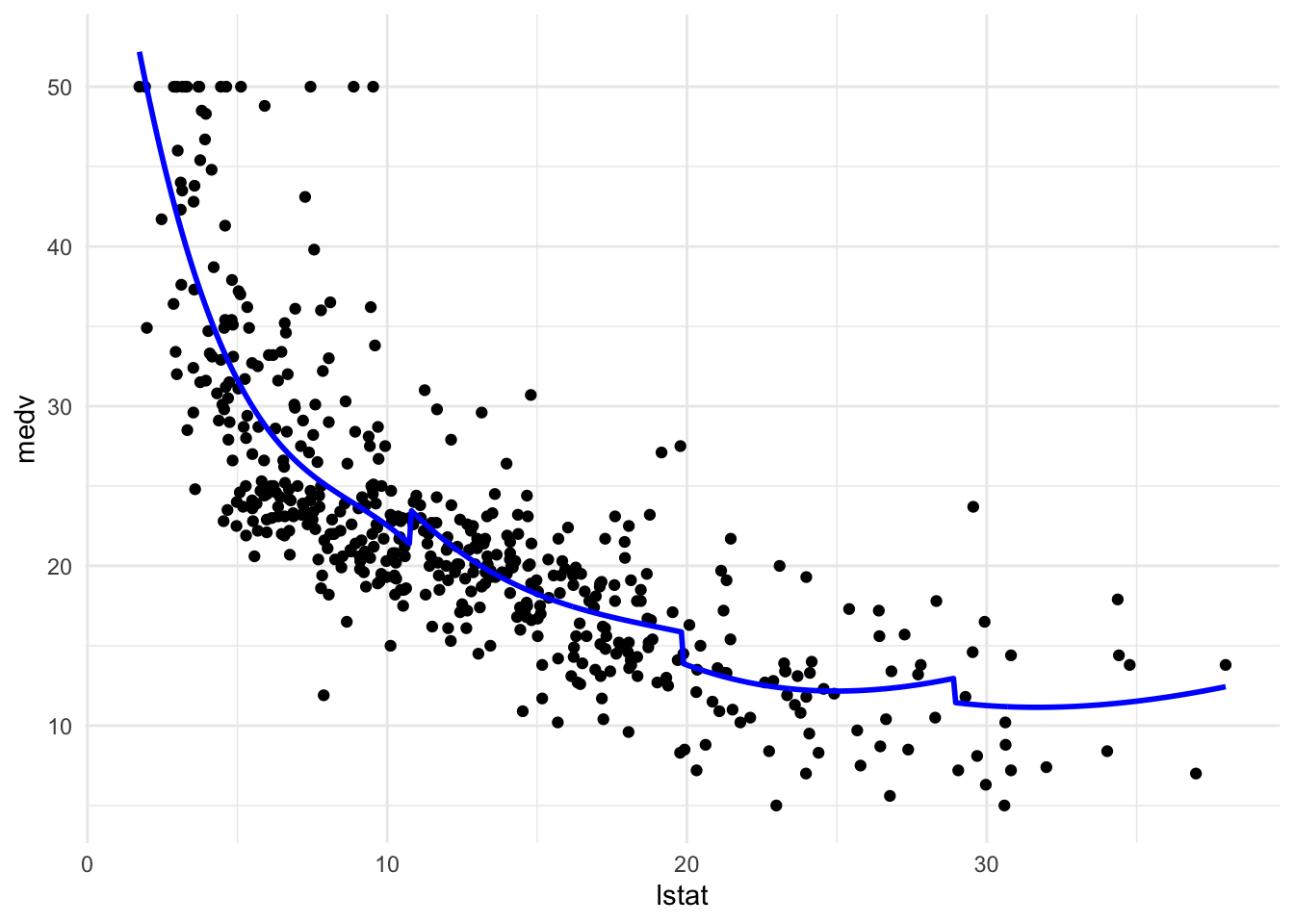
Piecewise polynomial regression
见https://dgoretzko.github.io/slv/practicals/08_nonlinear_regression/08_nonlinear_regression_answers.html
Splines
#dataframe #splines #ifelse
#create data frame from dataset
boston_tpb <- Boston %>% as_tibble %>% select(medv, lstat)
#added squared lstat and cubed lstat
boston_tpb <- boston_tpb %>% mutate(lstat2 = lstat^2, lstat3 = lstat^3)
#add a column lstat_tpb which is 0 below the median and has value (lstat - median(lstat))^3 above the median.
#ifelse的语法:(condition, true_value, false_value)
boston_tpb <- boston_tpb %>%
mutate(lstat_tpb = ifelse(lstat > median(lstat), (lstat - median(lstat))^3, 0))
#cubicsplinemodel #naturalcubicsplinemodel
#Create a linear model tpb_mod using the lm() function
tpb_mod <- lm(medv ~ lstat + lstat2 + lstat3 + lstat_tpb, data = boston_tpb)
summary(tpb_mod)
#Create a cubic spline model bs1_mod with a knot at the median using the bs() function.
bs1_mod <- lm(medv ~ bs(lstat, knots = median(lstat)), data = Boston)
summary(bs1_mod)
#plot
#Create a prediction plot from the bs1_mod object using the plot_pred() function.
pred_plot(bs1_mod)
#Create a natural cubic spline model (ns3_mod) with 3 degrees of freedom using the ns() function.
ns3_mod <- lm(medv ~ ns(lstat, df = 3), data = Boston)
pred_plot(ns3_mod)
#比较多个plot
library(cowplot)
plot_grid(
pred_plot(lin_mod) + ggtitle("Linear regression"),
pred_plot(pn3_mod) + ggtitle("Polynomial"),
pred_plot(pw5_mod) + ggtitle("Piecewise constant"),
pred_plot(pc3_mod) + ggtitle("Piecewise cubic"),
pred_plot(bs1_mod) + ggtitle("Cubic spline"),
pred_plot(ns3_mod) + ggtitle("Natural spline")
)
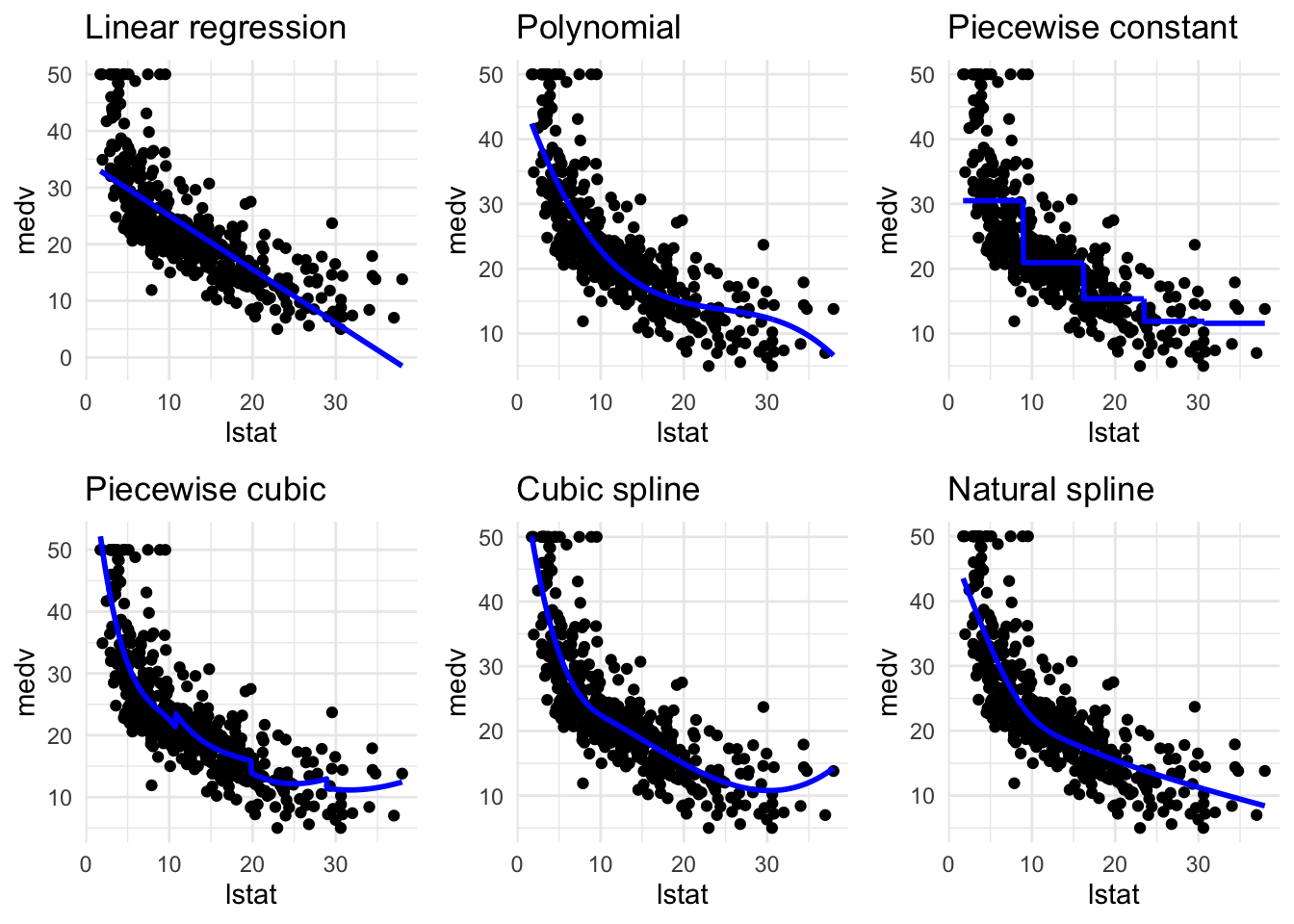
通过计算MSE评价各个模型的表现:
mse <- function(y_true, y_pred) mean((y_true - y_pred)^2)
# add a 12 split column to the boston dataset so we can cross-validate
boston_cv <- Boston %>% mutate(split = sample(rep(1:12, length.out = nrow(Boston))))
# prepare an output matrix with 12 slots per method for mse values
output_matrix <- matrix(nrow = 12, ncol = 6)
colnames(output_matrix) <- c("lin", "pn3", "pw5", "pc3", "bs1", "ns3")
# loop over the splits, run each method, and return the mse values
for (i in 1:12) {
train <- boston_cv %>% filter(split != i)
test <- boston_cv %>% filter(split == i)
brks <- c(-Inf, 7, 15, 22, Inf)
lin_mod <- lm(medv ~ lstat, data = train)
pn3_mod <- lm(medv ~ poly(lstat, 3), data = train)
pw5_mod <- lm(medv ~ cut(lstat, brks), data = train)
pc3_mod <- lm(medv ~ piecewise_cubic_basis(lstat, 3), data = train)
bs1_mod <- lm(medv ~ bs(lstat, knots = median(lstat)), data = train)
ns3_mod <- lm(medv ~ ns(lstat, df = 3), data = train)
output_matrix[i, ] <- c(
mse(test$medv, predict(lin_mod, newdata = test)),
mse(test$medv, predict(pn3_mod, newdata = test)),
mse(test$medv, predict(pw5_mod, newdata = test)),
mse(test$medv, predict(pc3_mod, newdata = test)),
mse(test$medv, predict(bs1_mod, newdata = test)),
mse(test$medv, predict(ns3_mod, newdata = test))
)
}
# this is the comparison of the methods
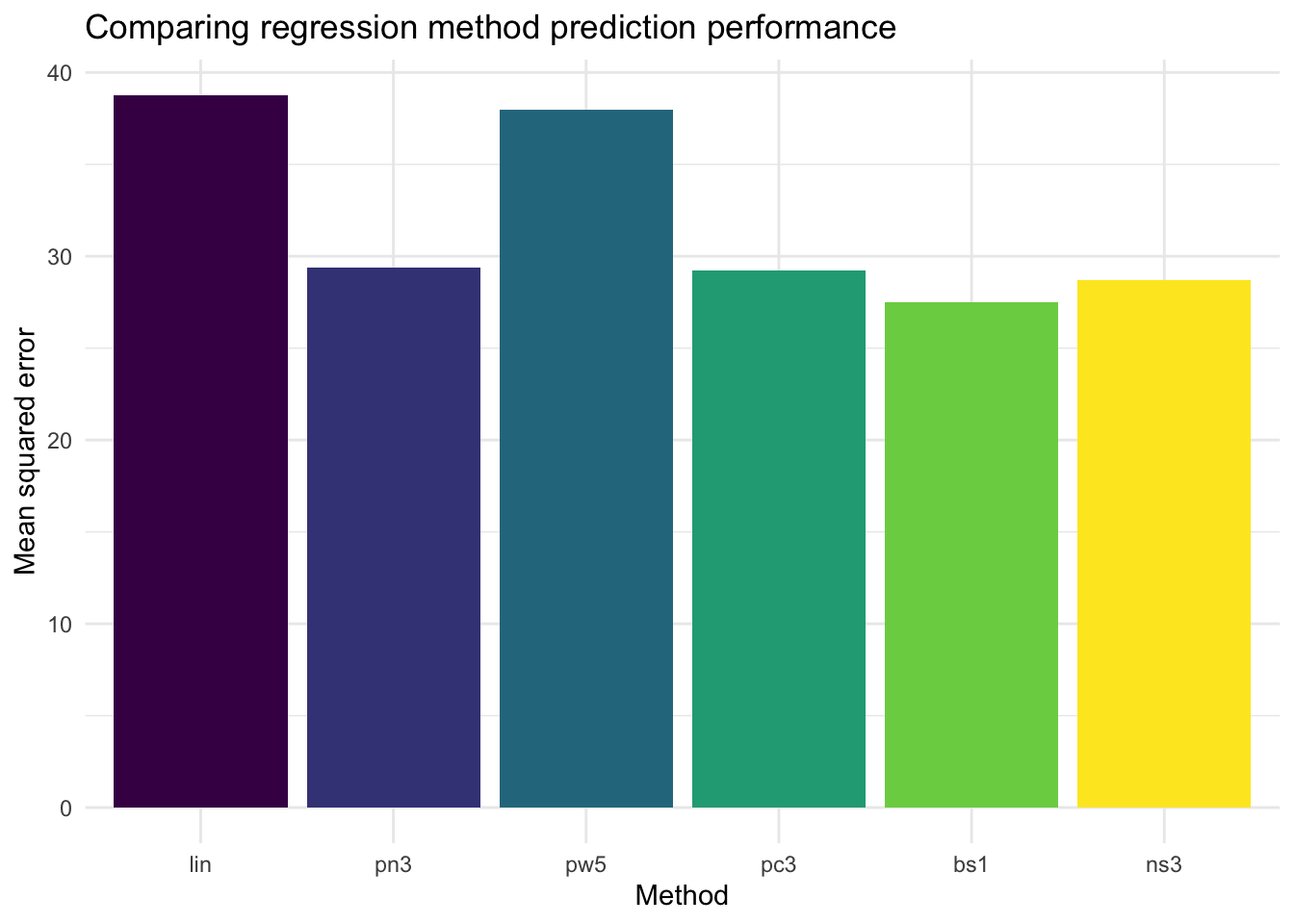
colMeans(output_matrix)
## lin pn3 pw5 pc3 bs1 ns3
## 38.76726 29.36707 37.95038 29.22791 27.51021 28.68858
# we can show it graphically too
tibble(names = as_factor(colnames(output_matrix)),
mse = colMeans(output_matrix)) %>%
ggplot(aes(x = names, y = mse, fill = names)) +
geom_bar(stat = "identity") +
theme_minimal() +
scale_fill_viridis_d(guide = "none") +
labs(
x = "Method",
y = "Mean squared error",
title = "Comparing regression method prediction performance"
)