3.8 Ensemble Methods - Bagging, RF, Boosting, Xgboost
#加载包(假设事先已经安装过包)
library(tidyverse)
library(magrittr)
library(psych)
library(caret)
library(gbm)
library(xgboost)
library(data.table)
library(ggforce)
#设定seed,读取数据
set.seed(45)
df <- readRDS("data/train_disease.RDS")
#基本EDA
head(df)
tail(df)
df %>%
select(-c(Gender, Disease)) %>%
describeBy(df$Disease, fast = TRUE)
#一些可视化(略)
#bagging
Bagging
#通过bagging预测outcome
cvcontrol <- trainControl(method = "repeatedcv",
number = 10,
allowParallel = TRUE)
bag_train <- train(Disease ~ .,
data = df,
method = 'treebag',
trControl = cvcontrol,
importance = TRUE)
#把各个变量的重要性可视化
bag_train %>%
varImp %>%
plot
#根据bagging结果生成confusion matrix,评价bagging的表现
confusionMatrix(predict(bag_train, type = "raw"),
df$Disease)
#但这个结果只是perfect training set performance而已
#这才是bagged model对out-of-sample数据的表现
bag_train
#randomforest #随机森林
Random Forest
rf_train <- train(Disease ~ .,
data = df,
method = 'rf',
trControl = cvcontrol,
importance = TRUE)
#可视化各个变量重要性
rf_train %>%
varImp %>%
plot
#生成confusion matrix和accuracy
rf_train
Boosting
gbm_train <- train(Disease ~ .,
data = df,
method = "gbm",
verbose = F,
trControl = cvcontrol)
#可视化各个variable的influence
summary(gbm_train)
#生成confusion matrix和accuracy
gbm_train
#github #xgboost #shap
Xgboost
#下载Github repository
library(devtools)
source_url("https://github.com/pablo14/shap-values/blob/master/shap.R?raw=TRUE")
#使用xgboost
train_x <- model.matrix(Disease ~ ., df)[,-1]
train_y <- as.numeric(df$Disease) - 1
xgboost_train <- xgboost(data = train_x,
label = train_y,
max.depth = 10,
eta = 1,
nthread = 4,
nrounds = 4,
objective = "binary:logistic",
verbose = 2)
pred <- tibble(Disease = predict(xgboost_train, newdata = train_x)) %>%
mutate(Disease = factor(ifelse(Disease < 0.5, 1, 2),
labels = c("Healthy", "Disease")))
table(pred$Disease, df$Disease)
#计算变量的shap rank score
shap_results <- shap.score.rank(xgboost_train,
X_train = train_x,
shap_approx = F)
#可视化各个variable的importance
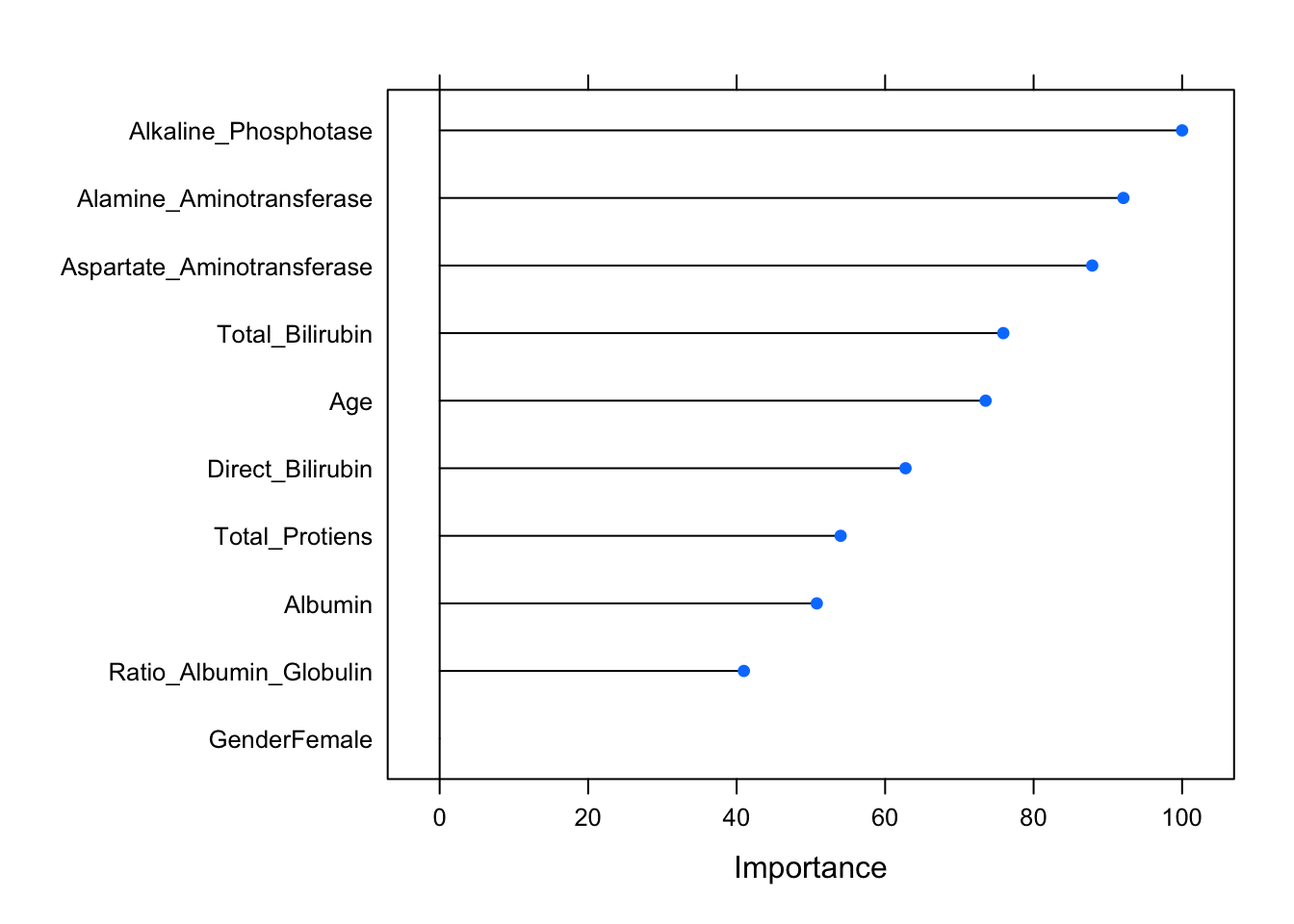
var_importance(shap_results)
#可视化每个feature每个individual的shap
shap_long <- shap.prep(shap = shap_results,
X_train = train_x)
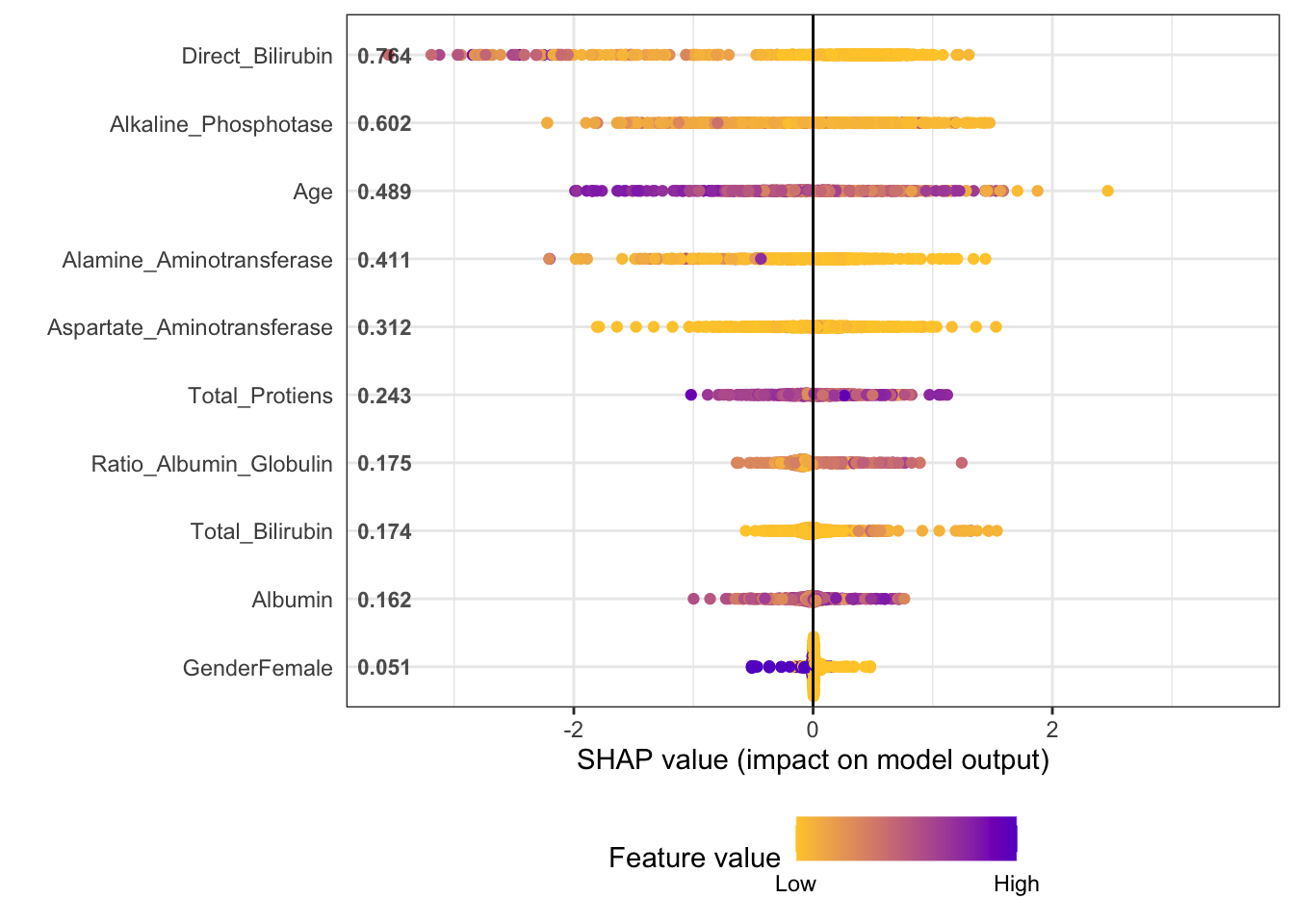
plot.shap.summary(shap_long)
xgb.plot.shap(train_x, features = colnames(train_x), model = xgboost_train, n_col = 3)
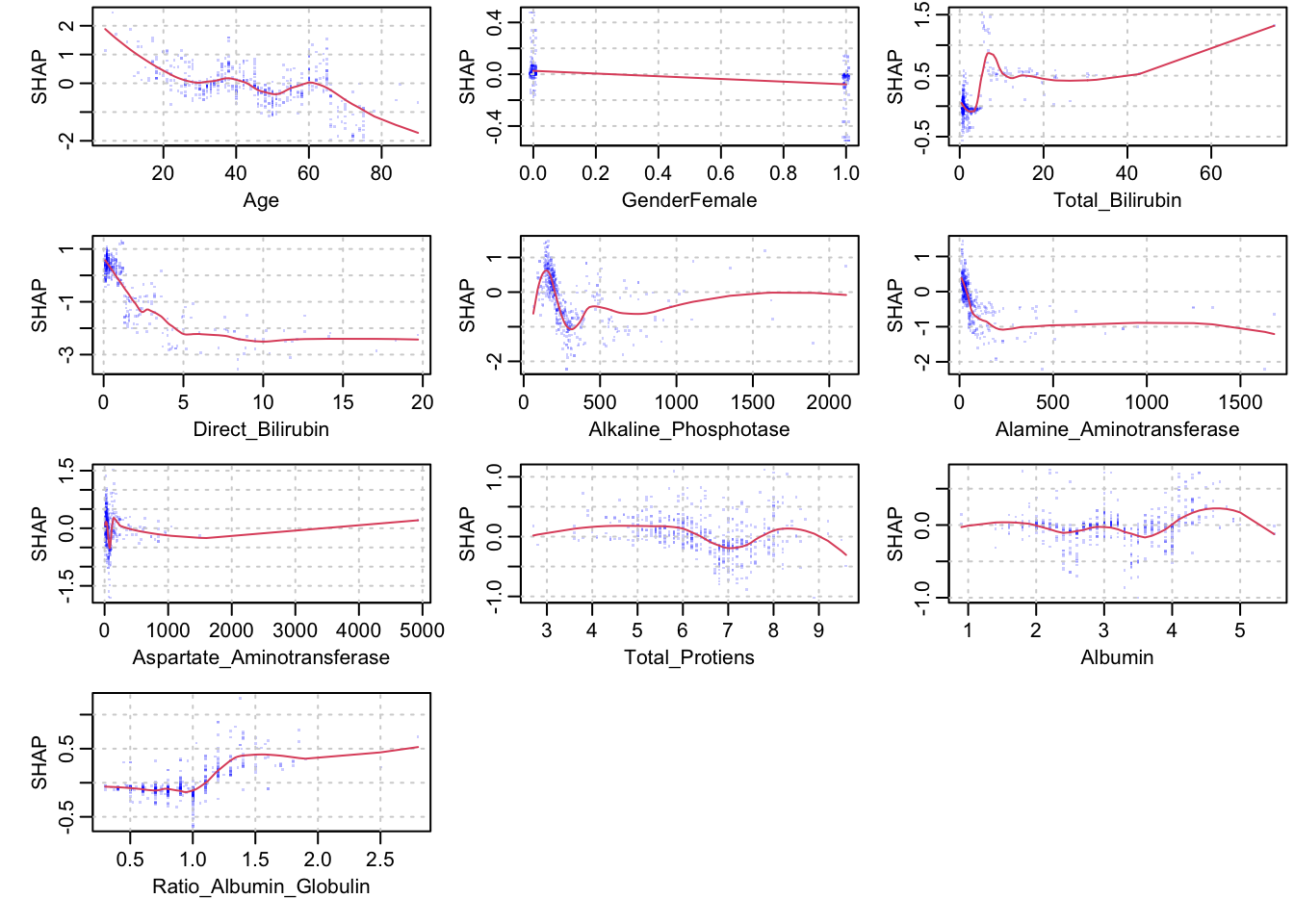
## The first plot shows, for example, that those with a high value for
## Direct_Bilirubin have a lower probability of being diseased. Also,
## Those with a higher age have a lower probability of being diseased,
## while those with a higher Albumin have a higher probability of being diseased.
## The second set of plots displays the marginal relationships of the SHAP values with the predictors. This conveys the same information, but in greater detail. The interpretability may be a bit tricky for the inexperienced data analyst.
#比较modelperformance
#比较哪个model表现最好
test <- readRDS("data/test_disease.RDS")
bag_test <- predict(bag_train, newdata = test)
rf_test <- predict(rf_train, newdata = test)
gbm_test <- predict(gbm_train, newdata = test)
xgb_test <- predict(xgboost_train, newdata = model.matrix(Disease ~ ., test)[,-1]) %>%
factor(x = ifelse(. < 0.5, 1, 2), levels = c(1,2), labels = c("Healthy", "Disease"))
list(bag_test,
rf_test,
gbm_test,
xgb_test) %>%
map(~ confusionMatrix(.x, test$Disease))